Projects
(Out of date)
Generalisable Subcellular Location Analysis
Most of subcellular location analysis in the past has been pursued using datasets that contain multiple copies of the same representative protein (i.e., a specific protein is selected to serve as a nuclear protein, another to serve as a microtubular protein).
Algorithms are then evaluated by measuring their ability to recognise this protein. It has then been inferred that the algorithm is thus capable of recognizing location patterns. Unfortunately, this is a leap of judgement that has not been tested. We tested this using a new dataset with multiple proteins standing in the same location group.
The algorithms we were using previously performed significantly worse in this new dataset and we thus employed new methods to achieve better results.
A manuscript describing this project and its results is in press (as of July 2013).
Unsupervised Subcellular Pattern Unmixing
The goal is to handle mixed subcellular location patterns (the result of having a protein or marker simmultaneously present in multiple organelles) in an unsupervised manner (without requiring that basic organelles be specified).
This was work with Tao Peng and Bob Murphy and the main result was that unsupervised unmixing could work as well as the supervised version.
More info on Unsupervised Unmixing
Nuclear Segmentation
The goal of this project was to quantitatively evaluate a series of nuclear segmentation algorithms for use in our work. We hand-segmented several images from two datasets and used those as gold standards to evaluate methods.
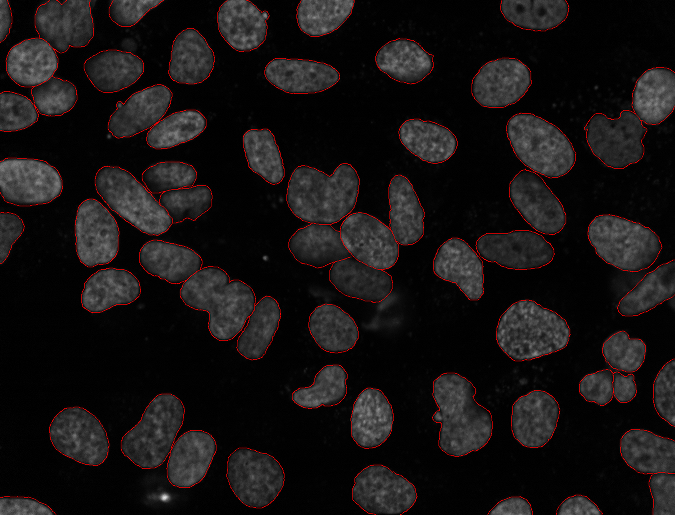
More info on Nuclear Segmentation
SLIF
SLIF is the [structured literature image finder]{.title-ref} (previously, [subcellular location image finder]{.title-ref}).
It searches through scientific journals, looking for images, which it categorises, makes available, and (in some cases, at least) analyses.
Programming for Scientists
Programming for Scientists is a course I designed for scientists who write code. The project grew out of informal discussions with others, but it crystalised around the observation that many scientists now write code as one of their primary activities (at least as defined on time spent) without a proper formal background for it.
Copyright (c) 2009-2023. Luis Pedro Coelho. All rights reserved.